Building AI-powered blood tests for autoimmune disease.
I’m cofounder and CEO of a diagnostics startup working to bring personalized medicine to autoimmune patients.
We spun out my Stanford PhD and postdoc research to develop a new sequencing and machine learning approach for clinical use.
Our work on reading the immune system’s records to track disease was published in Science and covered by The New York Times, The Wall Street Journal, STAT, Nature, GenomeWeb, and Eric Topol.
Previously, I was a product manager at Butterfly Network, where I helped make medical imaging accessible with an iPhone-connected ultrasound device now used in dozens of countries. I'm a software engineer by training — you can find my open-source projects on GitHub and, if you dig far enough, a decade of Stack Overflow answers I'd probably write differently today.
Interested in collaborating or joining our team?
We're based in NYC and work with clinicians and scientists across the US.
WHAT I'M BUILDING
Reading the immune system's disease sensors
The immune system keeps detailed records of disease exposures. But clinical diagnostics barely use it. We're changing that.
My team at Stanford, now at our spinoff, develops machine learning-powered blood tests to decode how B cells and T cells reconfigure their pattern-matching receptors when we get sick. Our proof of concept data shows these signals can help reliably distinguish diseases, including complex autoimmune conditions that are hard to diagnose today.
PREVIOUS WORK
Ultrasound on iPhone
Butterfly iQ, 2018 – 2019
I was the product manager responsible for Butterfly’s mobile ultrasound imaging software — determining what we build and how we build it. I reported directly to the head of product.
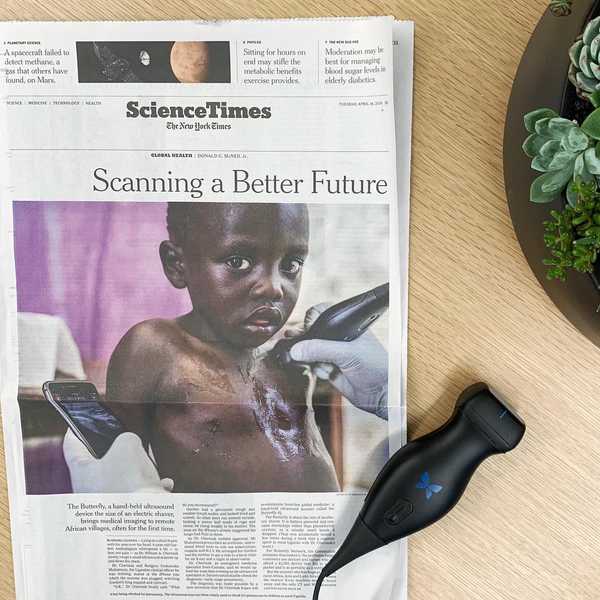
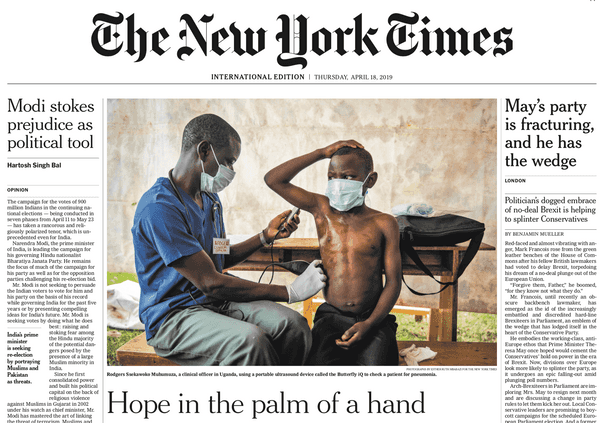
Butterfly iQ is the world’s first whole-body imager. iQ fits in your pocket, connects to your iPhone, and lets you see inside the body, for $2000 — a fraction of the price of a traditional ultrasound machine.
Butterfly’s mission is to make medical imaging more accessible, affordable, and intuitive. In the US, ultrasound is becoming a pillar of the physical examination. As for abroad, two thirds of the world has no access to even basic medical imaging. Handheld, simple-to-use ultrasound at a $2000 price point can help.
What makes Butterfly unique is that the piezoelectric crystals typically used in ultrasound have been replaced by custom-designed semiconductor chips, controlled entirely in software. Rather than swapping ultrasound probes to scan different parts of the body, a clinician scans with a single iQ probe and simply adjusts the scanning preset on their iPhone.
iQ is a medical device with the paradigms that make an iPhone app, an iPhone app. Connect your iQ and immediately start scanning. Swipe on the screen to adjust your image. Easily save to cloud storage and share with colleagues.

I was responsible for Butterfly’s iPhone and iPad software, which programs the iQ device, reconstructs images from ultrasound data, uploads captured images and videos to Butterfly secure cloud storage, and enables sharing and commenting for groups of physicians.
I managed the iQ software product from several months before launch to approximately one year after launch. I shipped:
- Core imaging features: the first Butterfly iQ app for iPhone and then for iPad; onboarding education revealed as the user learns the iQ scanning workflow; improved capture and annotation tools; and advanced imaging modes.
- Sales and marketing initiatives: a referral program that generated hundreds of thousands in revenue without a dime of incentives; safe deidentified image sharing; and the foundation for SaaS subscription management.
- Underlying infrastructure: speedy uploads, hardware-software compatibility, automatic diagnostic health checks, and lots and lots of usability refinements to get the iQ experience just right.
We won an Apple Design Award ("the Oscars of app development"), the SXSW Best of Show and Best in Health, Med, and Biotech awards, and other awards during my tenure.
Butterfly iQ was featured on the cover of the International New York Times, on the cover of the Science Times, and elsewhere.
I was also responsible for our software testing and release process along with my engineering partners, as we maintained a high-velocity release cadence, rare in medical devices.
PREVIOUS WORK
Remote ultrasound
Butterfly Tele-Guidance, 2018
Before I managed the core iQ product, my first assignment at Butterfly was to conceive of and build a way to empower non-ultrasound-trained clinicians with ultrasound insights for the first time, using telemedicine.
As a software engineer and product manager, I specced and built Butterfly Tele-Guidance, a green-field project combining ultrasound imaging with streaming video, computer vision, and augmented reality.
Imagine if any medical professional could unlock answers with ultrasound. Our technology emulates the feeling of an expert over your shoulder, guiding your hand as you scan. See press and footage from CES 2019.
Ultrasound is a game of millimeters. With Tele-Guidance, a novice scanner wouldn't need careful training to understand expert ultrasound terminology for maneuvering the probe. Instead, Tele-Guidance tracks the iQ probe’s position in the novice scanner’s iPhone camera feed. The remote expert can command precise adjustments in augmented reality, leading the novice scanner to the right image and insight.
Within my first 10 weeks, I built a fully functional iOS and web prototype to showcase at a national ultrasound trade show. Then I progressively refined the product through methodical user testing.
I worked as a solo engineer and product manager, with the help of a part-time product manager, pointers from several ultrasound software designers and engineers across the company, and guidance from the head of product and the chief medical officer.
Notable technologies used: Python, Typescript, React, WebRTC, Swift, SceneKit, Three.js, ArUco.
I ran the process to hire and onboard a tech lead to take over my day-to-day responsibilities, before I moved on to the core Butterfly iQ product.
PERSONAL
Music
Here's a jazz piano performance from a couple years ago with my band at Stanford.
- About
- Immune diagnostics
- Ultrasound on iPhone
- Remote ultrasound
- Music